15.5: Ribosomes na Protini awali
- Page ID
- 176493
Ujuzi wa Kuendeleza
- Eleza hatua tofauti katika awali ya protini
- Jadili jukumu la ribosomes katika awali ya protini
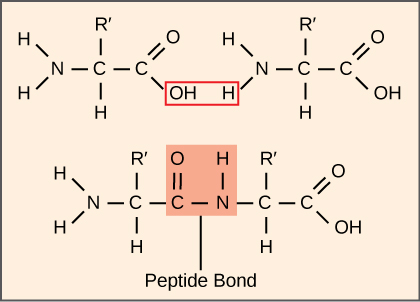
Awali ya protini hutumia nishati zaidi ya seli kuliko mchakato wowote wa metabolic. Kwa upande mwingine, protini akaunti kwa wingi zaidi kuliko sehemu nyingine yoyote ya viumbe hai (isipokuwa maji), na protini hufanya karibu kila kazi ya seli. Mchakato wa tafsiri, au awali ya protini, unahusisha uamuzi wa ujumbe wa mRNA kwenye bidhaa za polipeptidi. Asidi amino hupigwa kwa pamoja kwa kuunganisha vifungo vya peptidi kwa urefu kuanzia takriban 50 mabaki ya asidi amino hadi zaidi ya 1,000. Kila asidi amino binafsi ina kundi la amino (NH 2) na kikundi cha carboxyl (COOH). Polypeptidi hutengenezwa wakati kundi la amino la asidi amino moja linaunda amidi (yaani, peptide) dhamana na kundi la carboxyl la asidi nyingine ya amino (Kielelezo\(\PageIndex{1}\)). Mmenyuko huu huchochewa na ribosomu na huzalisha molekuli moja ya maji.
Protini awali Mashine
Mbali na template ya mRNA, molekuli nyingi na macromolecules huchangia mchakato wa tafsiri. Utungaji wa kila sehemu unaweza kutofautiana kwa aina mbalimbali; kwa mfano, ribosomu zinaweza kuwa na idadi tofauti za RRNAs na polipeptidi kulingana na viumbe. Hata hivyo, miundo ya jumla na kazi za mashine ya awali ya protini zinafanana na bakteria hadi seli za binadamu. Tafsiri inahitaji pembejeo ya template ya mRNA, ribosomes, trNAs, na mambo mbalimbali ya enzymatic.
Unganisha na Kujifunza

Bonyeza kupitia hatua ya PBS hii maingiliano kuona protini awali katika hatua.
Ribosomu
Hata kabla ya mRNA kutafsiriwa, kiini lazima kuwekeza nishati ya kujenga kila moja ya ribosomu yake. Katika E. koli, kuna ribosomu kati ya 10,000 na 70,000 zilizopo katika kila seli wakati wowote. Ribosome ni macromolecule tata linajumuisha RNAs miundo na kichocheo, na polypeptides nyingi tofauti. Katika eukaryotes, nucleolus ni maalumu kabisa kwa ajili ya awali na mkutano wa RRNAs.
Ribosomu zipo katika cytoplasm katika prokaryotes na katika cytoplasm na reticulum mbaya endoplasmic katika eukaryotes. Mitochondria na kloroplasti pia zina ribosomu zao wenyewe katika tumbo na stroma, ambazo zinafanana zaidi na ribosomu za prokaryotiki (na zina uelewa sawa wa madawa ya kulevya) kuliko ribosomu nje ya utando wao wa nje katika saitoplazimu. Ribosomes hutenganisha katika subunits kubwa na ndogo wakati wao si synthesizing protini na reassociate wakati wa kuanzishwa kwa tafsiri. Katika E. coli, subunit ndogo inaelezewa kama 30S, na subunit kubwa ni 50S, kwa jumla ya 70S (kumbuka kwamba vitengo vya Svedberg sio nyongeza). Ribosomu za mamalia zina subunit ndogo ya 40S na subunit kubwa ya 60, kwa jumla ya 80s. Subunit ndogo ni wajibu wa kumfunga template ya mRNA, wakati subunit kubwa hufunga trNAs kwa sequentially. Kila molekuli ya mRNA hutafsiriwa wakati huo huo na ribosomu nyingi, protini zote za kuunganisha katika mwelekeo huo: kusoma mRNA kutoka 5' hadi 3' na kuunganisha polipeptidi kutoka kwa mwisho wa N hadi kwenye terminal C. Muundo kamili wa MRNA/Poly-ribosome huitwa polysome.
RNAs
RNAs ni molekuli za RNA za kimuundo zilizoandikwa kutoka jeni na RNA polymerase III. Kulingana na aina, aina 40 hadi 60 za trNAs zipo katika cytoplasm. Kutumikia kama adapta, TRNAs maalum hufunga kwa utaratibu kwenye template ya mRNA na kuongeza asidi amino inayofanana na mnyororo wa polipeptidi. Kwa hiyo, RNAs ni molekuli ambazo kwa kweli “hutafsiri” lugha ya RNA katika lugha ya protini.
Kati ya 64 inawezekana mRNA kodoni-au mchanganyiko triplet ya A, U, G, na C—tatu kutaja kusitishwa kwa protini awali na 61 kutaja nyongeza ya amino asidi kwa mnyororo polipeptidi. Kati ya hizi 61, codon moja (Agosti) pia inajumuisha uanzishwaji wa tafsiri. Kila anticodon ya tRNA inaweza msingi jozi na moja ya codons mRNA na kuongeza asidi amino au kusitisha tafsiri, kulingana na kanuni za maumbile. Kwa mfano, kama mlolongo CUA ilitokea kwenye template mRNA katika sura sahihi ya kusoma, ingekuwa kumfunga tRNA kuonyesha mlolongo nyongeza, GAU, ambayo itakuwa wanaohusishwa na leucine amino asidi.
Kama molekuli ya adapta ya tafsiri, ni ajabu kwamba trNAs zinaweza kufaa maalum sana katika mfuko mdogo kama huo. Fikiria kwamba TRNAs zinahitaji kuingiliana na mambo matatu: 1) zinapaswa kutambuliwa na synthetase sahihi ya aminoacyl (angalia hapa chini); 2) wanapaswa kutambuliwa na ribosomes; na 3) wanapaswa kumfunga mlolongo sahihi katika mRNA.
Synthetases ya Aminoacyl RNA
Mchakato wa awali kabla ya TRNA na RNA polymerase III hujenga tu sehemu ya RNA ya molekuli ya adapta. Asidi ya amino inayofanana lazima iongezwe baadaye, mara moja tRNA inachukuliwa na kusafirishwa kwa cytoplasm. Kupitia mchakato wa tRNA “kumshutumu,” kila molekuli ya tRNA inaunganishwa na asidi amino yake sahihi na kundi la enzymes inayoitwa aminoacyl trNA synthetases. Angalau aina moja ya aminoacyl trNA synthetase ipo kwa kila moja ya asidi amino 20; idadi halisi ya synthetases aminoacyl tRNA inatofautiana na spishi. Enzymes hizi hufunga kwanza na hidrolyze ATP ili kuchochea dhamana ya juu-nishati kati ya asidi amino na monophosphate ya adenosini (AMP); molekuli ya pyrophosphate inafukuzwa katika mmenyuko huu. Asidi ya amino iliyoamilishwa huhamishiwa kwenye tRNA, na AMP inatolewa.
Mfumo wa Protini awali
Kama ilivyo na awali ya mRNA, awali ya protini inaweza kugawanywa katika awamu tatu: uanzishwaji, upungufu, na kukomesha. Mchakato wa tafsiri ni sawa katika prokaryotes na eukaryotes. Hapa tutaweza kuchunguza jinsi tafsiri hutokea katika E. coli, mwakilishi prokaryote, na kutaja tofauti yoyote kati ya tafsiri prokaryotic na eukaryotic.
Uanzishwaji wa Tafsiri
Protini awali huanza na malezi ya tata ya kuanzishwa. Katika E. coli, tata hii inahusisha ribosomu ndogo ya 30S, template ya mRNA, mambo matatu ya uanzishwaji (IFs; IF-1, IF-2, na IF-3), na mwanzilishi maalum wa RNA, inayoitwa\(\text{tRNA}_\text{f}^\text{Met}\). The initiator tRNA interacts with the start codon AUG (or rarely, GUG), links to a formylated methionine called fMet, and can also bind IF-2. Formylated methionine is inserted by \(\text{fMet} - \text{tRNA}_\text{f}^\text{Met}\) at the beginning of every polypeptide chain synthesized by E. coli, but it is usually clipped off after translation is complete. When an in-frame AUG is encountered during translation elongation, a non-formylated methionine is inserted by a regular Met-tRNAMet.
In E. coli mRNA, a sequence upstream of the first AUG codon, called the Shine-Dalgarno sequence (AGGAGG), interacts with the rRNA molecules that compose the ribosome. This interaction anchors the 30S ribosomal subunit at the correct location on the mRNA template. Guanosine triphosphate (GTP), which is a purine nucleotide triphosphate, acts as an energy source during translation—both at the start of elongation and during the ribosome’s translocation.
In eukaryotes, a similar initiation complex forms, comprising mRNA, the 40S small ribosomal subunit, IFs, and nucleoside triphosphates (GTP and ATP). The charged initiator tRNA, called Met-tRNAi, does not bind fMet in eukaryotes, but is distinct from other Met-tRNAs in that it can bind IFs.
Instead of depositing at the Shine-Dalgarno sequence, the eukaryotic initiation complex recognizes the 7-methylguanosine cap at the 5' end of the mRNA. A cap-binding protein (CBP) and several other IFs assist the movement of the ribosome to the 5' cap. Once at the cap, the initiation complex tracks along the mRNA in the 5' to 3' direction, searching for the AUG start codon. Many eukaryotic mRNAs are translated from the first AUG, but this is not always the case. According to Kozak’s rules, the nucleotides around the AUG indicate whether it is the correct start codon. Kozak’s rules state that the following consensus sequence must appear around the AUG of vertebrate genes: 5'-gccRccAUGG-3'. The R (for purine) indicates a site that can be either A or G, but cannot be C or U. Essentially, the closer the sequence is to this consensus, the higher the efficiency of translation.
Once the appropriate AUG is identified, the other proteins and CBP dissociate, and the 60S subunit binds to the complex of Met-tRNAi, mRNA, and the 40S subunit. This step completes the initiation of translation in eukaryotes.
Translation, Elongation, and Termination
In prokaryotes and eukaryotes, the basics of elongation are the same, so we will review elongation from the perspective of E. coli. The 50S ribosomal subunit of E. coli consists of three compartments: the A (aminoacyl) site binds incoming charged aminoacyl tRNAs. The P (peptidyl) site binds charged tRNAs carrying amino acids that have formed peptide bonds with the growing polypeptide chain but have not yet dissociated from their corresponding tRNA. The E (exit) site releases dissociated tRNAs so that they can be recharged with free amino acids. There is one exception to this assembly line of tRNAs: in E. coli, \(\text{fMet} - \text{tRNA}_\text{f}^\text{Met}\) is capable of entering the P site directly without first entering the A site. Similarly, the eukaryotic Met-tRNAi, with help from other proteins of the initiation complex, binds directly to the P site. In both cases, this creates an initiation complex with a free A site ready to accept the tRNA corresponding to the first codon after the AUG.
During translation elongation, the mRNA template provides specificity. As the ribosome moves along the mRNA, each mRNA codon comes into register, and specific binding with the corresponding charged tRNA anticodon is ensured. If mRNA were not present in the elongation complex, the ribosome would bind tRNAs nonspecifically.
Elongation proceeds with charged tRNAs entering the A site and then shifting to the P site followed by the E site with each single-codon “step” of the ribosome. Ribosomal steps are induced by conformational changes that advance the ribosome by three bases in the 3' direction. The energy for each step of the ribosome is donated by an elongation factor that hydrolyzes GTP. Peptide bonds form between the amino group of the amino acid attached to the A-site tRNA and the carboxyl group of the amino acid attached to the P-site tRNA. The formation of each peptide bond is catalyzed by peptidyl transferase, an RNA-based enzyme that is integrated into the 50S ribosomal subunit. The energy for each peptide bond formation is derived from GTP hydrolysis, which is catalyzed by a separate elongation factor. The amino acid bound to the P-site tRNA is also linked to the growing polypeptide chain. As the ribosome steps across the mRNA, the former P-site tRNA enters the E site, detaches from the amino acid, and is expelled (Figure \(\PageIndex{2}\)). Amazingly, the E. coli translation apparatus takes only 0.05 seconds to add each amino acid, meaning that a 200-amino acid protein can be translated in just 10 seconds.
Art Connection
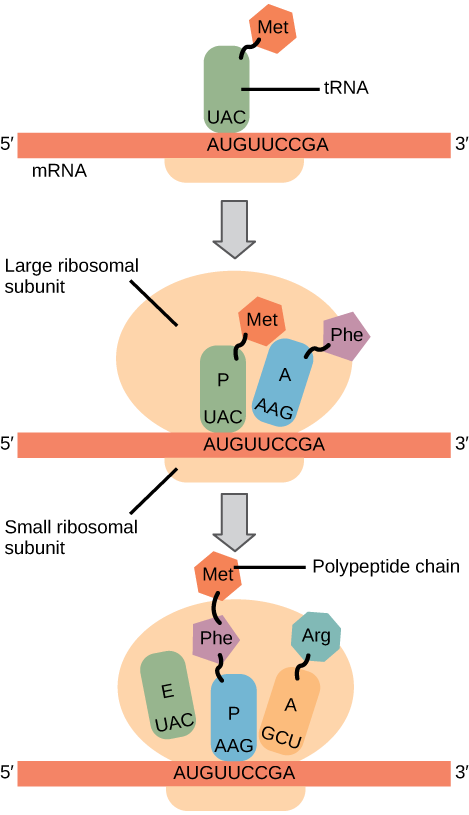
Many antibiotics inhibit bacterial protein synthesis. For example, tetracycline blocks the A site on the bacterial ribosome, and chloramphenicol blocks peptidyl transfer. What specific effect would you expect each of these antibiotics to have on protein synthesis?
Tetracycline would directly affect:
- tRNA binding to the ribosome
- ribosome assembly
- growth of the protein chain
Chloramphenicol would directly affect
- tRNA binding to the ribosome
- ribosome assembly
- growth of the protein chain
Termination of translation occurs when a nonsense codon (UAA, UAG, or UGA) is encountered. Upon aligning with the A site, these nonsense codons are recognized by release factors in prokaryotes and eukaryotes that instruct peptidyl transferase to add a water molecule to the carboxyl end of the P-site amino acid. This reaction forces the P-site amino acid to detach from its tRNA, and the newly made protein is released. The small and large ribosomal subunits dissociate from the mRNA and from each other; they are recruited almost immediately into another translation initiation complex. After many ribosomes have completed translation, the mRNA is degraded so the nucleotides can be reused in another transcription reaction.
Protein Folding, Modification, and Targeting
During and after translation, individual amino acids may be chemically modified, signal sequences may be appended, and the new protein “folds” into a distinct three-dimensional structure as a result of intramolecular interactions. A signal sequence is a short tail of amino acids that directs a protein to a specific cellular compartment. These sequences at the amino end or the carboxyl end of the protein can be thought of as the protein’s “train ticket” to its ultimate destination. Other cellular factors recognize each signal sequence and help transport the protein from the cytoplasm to its correct compartment. For instance, a specific sequence at the amino terminus will direct a protein to the mitochondria or chloroplasts (in plants). Once the protein reaches its cellular destination, the signal sequence is usually clipped off.
Many proteins fold spontaneously, but some proteins require helper molecules, called chaperones, to prevent them from aggregating during the complicated process of folding. Even if a protein is properly specified by its corresponding mRNA, it could take on a completely dysfunctional shape if abnormal temperature or pH conditions prevent it from folding correctly.
Summary
The players in translation include the mRNA template, ribosomes, tRNAs, and various enzymatic factors. The small ribosomal subunit forms on the mRNA template either at the Shine-Dalgarno sequence (prokaryotes) or the 5' cap (eukaryotes). Translation begins at the initiating AUG on the mRNA, specifying methionine. The formation of peptide bonds occurs between sequential amino acids specified by the mRNA template according to the genetic code. Charged tRNAs enter the ribosomal A site, and their amino acid bonds with the amino acid at the P site. The entire mRNA is translated in three-nucleotide “steps” of the ribosome. When a nonsense codon is encountered, a release factor binds and dissociates the components and frees the new protein. Folding of the protein occurs during and after translation.
Art Connections
Figure \(\PageIndex{2}\): Many antibiotics inhibit bacterial protein synthesis. For example, tetracycline blocks the A site on the bacterial ribosome, and chloramphenicol blocks peptidyl transfer. What specific effect would you expect each of these antibiotics to have on protein synthesis?
Tetracycline would directly affect:
- tRNA binding to the ribosome
- ribosome assembly
- growth of the protein chain
Chloramphenicol would directly affect
- tRNA binding to the ribosome
- ribosome assembly
- growth of the protein chain
- Answer
-
Tetracycline: a; Chloramphenicol: c.
Glossary
- aminoacyl tRNA synthetase
- enzyme that “charges” tRNA molecules by catalyzing a bond between the tRNA and a corresponding amino acid
- initiator tRNA
- in prokaryotes, called \(\text{tRNA}_\text{f}^\text{Met}\); in eukaryotes, called tRNAi; a tRNA that interacts with a start codon, binds directly to the ribosome P site, and links to a special methionine to begin a polypeptide chain
- Kozak’s rules
- determines the correct initiation AUG in a eukaryotic mRNA; the following consensus sequence must appear around the AUG: 5’-GCC(purine)CCAUGG-3’; the bolded bases are most important
- peptidyl transferase
- RNA-based enzyme that is integrated into the 50S ribosomal subunit and catalyzes the formation of peptide bonds
- polysome
- mRNA molecule simultaneously being translated by many ribosomes all going in the same direction
- Shine-Dalgarno sequence
- (AGGAGG); initiates prokaryotic translation by interacting with rRNA molecules comprising the 30S ribosome
- signal sequence
- short tail of amino acids that directs a protein to a specific cellular compartment
- start codon
- AUG (or rarely, GUG) on an mRNA from which translation begins; always specifies methionine


